Many genetic variants are novel or rare which makes difficult their clinical interpretation. The DECIPHER Consortium was initiated in 2004 as a community of academic centers of Clinical Genetics who submit consented, anonymized genotype and phenotype data from patients with rare genomic disorders for sharing with other clinicians and researchers. The identification of patients sharing variants in a given locus with common phenotypic features leads to greater certainty in the clinical interpretation of these variants. As of January 6, 2015, there are 18 539 publicly available patient record, 51 496 phenotype observation in these patients, and 27 175 publicly available copy-number variants in this database.
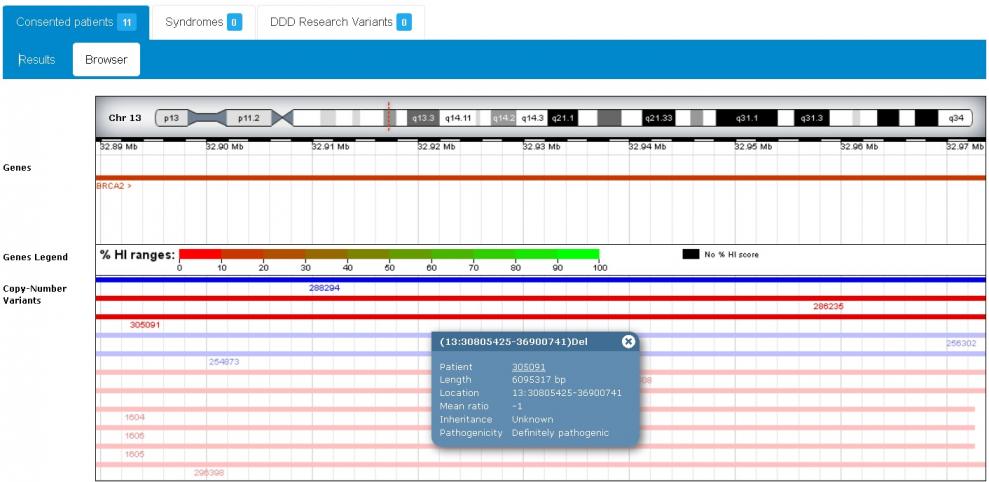
DECIPHER can be search by phenotype, by genomic position, band, gene, pathogenicity, variant consequence, etc. Results are presented as a table or can be visualized in a browser. This browser contains different tracks where variants can be visualized in the context of other data.
Learn more on DECIPHER and how to use it to make sense of genetic variants at the workshop “Making Sense of Variation”.
You can also contact Rolando Garcia-Milian with questions on this or any other variation tool,
References
DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources. Firth, H.V. et al (2009). Am.J.Hum.Genet 84, 524-533 (DOI: dx.doi.org/10/1016/j.ajhg.2009.03.010)