The Yale Medical Library is providing to all Yale affiliates free access to two of the most powerful commercial bioinformatics tools for the analysis of omics data: MetaCore and Ingenuity Pathway Analysis. This is part of a pilot project conducted by the medical library in order to find sustainable and long term access to these tools. Please register for these upcoming trainings if you are interested in learning how to use these tools or if you need a refresher.
For questions on how to register for an account or comments please contact Rolando Milian
Title: Introduction to Ingenuity Pathway Analysis
Description:
- What is IPA and what questions can it address?
- Overview of key features in IPA
- Ingenuity Knowledge Base
- Search & Pathway Building - Gene/ Chemical, Functions, Drug Targets
- Advanced Search: Limiting results to a molecule type, family or disease-association.
- Building pathways: Creating a pathway, pathway navigating, Using Build and Overlay tools
- Bioprofiler
- Dataset Analysis: Interpretation of Gene, Transcript, Protein and Metabolite Data
- Data Upload and Analysis: Uploading and formatting a dataset, setting analysis parameters and running an analysis
- Pathway Analysis and Canonical Pathways
- Downstream Effects Analysis and identifying downstream functions and processes that are likely affected
- Upstream regulators Analysis
- Causal Network Analysis and identifying likely root regulators
- Regulator Effects Analysis to link upstream regulators with downstream functions and processes that are affected
- Comparison analysis and comparing multiple observations
Date & Time: 9:00am - 12:00pm, Tuesday, October 27, 2015
Location: H-203, Jane Ellen Hope Building, 315 Cedar St, New Haven CT
Presenter: Field Scientist QIAGEN Informatics
Title: MetaCore: Getting the most from your "omics" analysis (Introductory session)
Description: The ability to generate massive amounts of data with "omics" analysis begs the need for a tool to analyze and prioritize the biological relevance of this information. GeneGo provides a solution for using "omics" gene lists to generate and prioritize hypotheses with MetaCore. This tutorial highlights how to work with different types of data (genomics, proteomics, metabolomics and interaction data) beginning with how to upload gene lists and expression data (if available). Here we demonstrate data manager capabilities including how to upload, batch upload, store, share and check data properties and signal distribution. We then focus on how MetaCore uses your gene list to extract functional relevance by determining the most enriched processes across several ontologies. This entails a detailed lesson on how to prioritize your hypothesis using the statistically significance enrichment histograms and associate highly interactive GeneGo Maps and pre-built networks. We further emphasize the role of expression data in your analysis and the ability to visually predict experimental results, associated disease and possible drug targets. Lastly we highlight the benefits of using MetaCore workflows to compare data sets and work with experiment intersections.
Date & Time: 10:00am - 12:00pm, Tuesday, November 3, 2015
Location: C-103 - SHM 333 Cedar St, New Haven CT 06520
Presenter: Dr. Matthew Wampole, Solution Scientist, IP & Science, Thomson Reuters
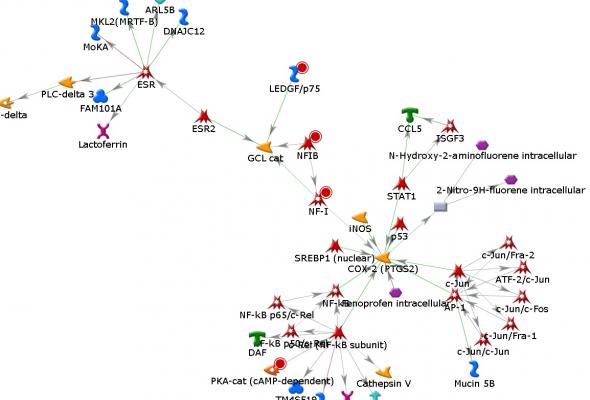
Title: MetaCore: Getting the most from your "omics" analysis (Advanced)
Description: In the advanced tutorial, we will explore uses of our network building algorithms and methods for hypothesizing key hubs passed on data. We will begin this session with a discussion on using the Key Pathway Advisor to hypothesize key hubs regulating gene expression data. The session will then review ways of using the 11 network building algorithms in MetaCore. The first example will review how to build a network purely from the curated knowledge within MetaCore. Then we will go through an example of using omics data to build a network of interactions to better understand the relationships within our data.
Date & Time: 1:00pm - 3:00pm, Tuesday, November 3, 2015
Location: C-103 - SHM 333 Cedar St, New Haven CT 06520
Presenter: Dr. Matthew Wampole, Solution Scientist, IP & Science, Thomson Reuters
Join the End-user Bioinformatics Group and become a member of a community that collaborates on end-user bioinformatics events, training sessions, resources, and tools that support biomedical research at Yale.